Isolation and identification of Cryptosporidium from water samples in Bangladesh
Volume 2, Issue 1, February 2015, Pages 118–124
Sharifuzzaman1, Md. Shahiduzzaman1![]()
![]() , Anisuzzaman1, Md. Shafiullah Parvez2, Antonio Barragan3,4
, Anisuzzaman1, Md. Shafiullah Parvez2, Antonio Barragan3,4
1Department of Parasitology, Bangladesh Agricultural University, Mymensingh-2202, Bangladesh
2Department of Microbiology and Hygiene, Bangladesh Agricultural University, Mymensingh-2202, Bangladesh
3Department of Molecular Biosciences, The Wenner-Gren Institute, Stockholm University, Stockholm, Sweden
4Center for Infectious Medicine, Department of Medicine, Karolinska Institutet, Stockholm, Sweden
| ABSTRACT | Get Full Text PDF |
Water-borne cryptosporidiosis, a neglected parasitic disease, presents a serious threat to human health due to the ubiquitous distribution of Cryptosporidium species affecting humans and animals, and due to the resistance of the oocysts to harsh environmental conditions, various disinfectants, and some common therapeutic agents. Water is a great reservoir for contamination and a potential medium of transmission of the pathogen. The present study was aimed to isolate and identify Cryptosporidium oocysts in water samples in Bangladesh. Water samples from small ponds, waste water bodies and drains were collected from different areas of the Mymensingh and Kushtia districts of Bangladesh. To detect Cryptosporidium, the samples were assessed by microscopy, using the conventional Ziehl-Neelsen staining method, and by PCR. By microscopy, Cryptosporidium oocysts were detected in 5 (25%) of 20 samples examined collected from the Mymensingh district, which were further confirmed by PCR. Importantly, Cryptosporidium was detected by PCR in drain water but was not detected by conventional microscopy. In the Kushtia district, 2 (20%) of the 10 samples examined by microscopy were positive. Cryptosporidium was not detected in waste water by conventional microscopy and PCR. However, the results confirm the presence of Cryptosporidium in different types of sources of water in both Mymensingh and Kushtia districts, demonstrating the existence of Cryptosporidium in the study areas. Further studies are need to explore the present status of prevalence of Cryptosporidium in humans and animals in order to better understand the transmission dynamics of the parasite and thereafter take necessary measures to control and/or prevent the disease.
Key words: Cryptosporidium, oocyst, water, Bangladesh.
![]() Corresponding author. Tel.: +8801716460558
Corresponding author. Tel.: +8801716460558
E-mail address: szaman@bau.edu.bd (M Shahiduzzaman)
How to cite this article: Sharifuzzaman, M Shahiduzzaman, Anisuzzaman, MS Parvez and A Barragan (2015). Isolation and identification of Cryptosporidium from water sample in Bangladesh. International Journal of Natural and Social Sciences, 2(1): 118-124.
INTRODUCTION
Cryptosporidium is a zoonotic protozoan parasite which infects the intestinal epithelium of diverse mammals including humans, and causes a diarrheal disease, Cryptosporidiosis (Tzipori, and Ward, 2002). Cryptosporidium is the most notorious one among more than 150 potentially waterborne pathogens (WHO, 2004), causes gastrointestinal illness in a variety of mammals. The infection is usually self-limiting in immune-competent individuals, but fatal in immuno-compromised individuals, e.g., acquired immune deficiency syndrome (AIDS) or leukaemia patients, taking immunosuppressive agents, malnourished children and elderly individuals (Current et al., 1983; Alves et al., 2001; Mohandas 2002). Cryptosporidiosis is prevalent worldwide (Dalle et al., 2003; Leoni et al., 2007; Lake et al., 2008; Zintl et al., 2008).
Water is the principal reservoir for contamination and potential medium of transmission of the parasite Cryptosporidium oocysts, which are excreted by infected animals and humans, are commonly found in lakes, rivers and streams that serve as drinking water sources. Cryptosporidium oocysts are resistant to normal environmental condition and common disinfectants. Traditional epidemiological investigations only provide information on the prevalence of this parasite but do not provide any information for tracking infection sources and or transmission dynamics of Cryptosporidium. Molecular epidemiological studies have significantly improved our knowledge of cryptosporidiosis. Recently, molecular diagnostic tools have been developed to assess the human infection potential of Cryptosporidium oocysts in water and to track the sources of contamination.
Cryptosporidium species are reported to be a significant cause of diarrhoeal illness of young children especially less than 5 years of age in Bangladesh (Rahman et al., 1990; Bhattacharya et al., 1997, Albert et al., 1999). About a decade ago, infection with Cryptosporidium species were reported in about 1.4 -8.4% of diarrhoeal patients (Haque et al., 2003; Khan et al., 2004) from International Centre for Diarrheal Disease Research, Bangladesh in Dhaka, (ICDDR’B). On the other hand, about three decades ago, the disease was also found prevalent among the animal population in Bangladesh (Rahman et al., 1985). Clinical cases in most of places in Bangladesh are treated as undiagnosed diarrhoeal patients. Despite enormous effect on animal and human health, presently very little attention has been paid to determine the present status of Cryptosporidium infection in Bangladesh.
The small subunit (SSU) rRNA genes of Cryptosporidium were usually targeted to detect and differentiate Cryptosporidium from animal and human sources (Xiao et al., 1999). The recent epidemiological data provided information on the infection potentials and transmission dynamics of Cryptosporidium in developed and developing countries. Bangladesh is one of the developing countries with poor water purification, supply, and waste management but Cryptosporidium is endemic, nonetheless, molecular epidemiological investigations are yet to be performed to determine reliable, solid and applicable information. The study was conducted to adopt a method for isolation and detection of Cryptosporidium from different water sources in Bangladesh.
MATERIALS AND METHOD
Study area and period
Water samples were collected from the Mymensingh and Kushtia districts of Bangladesh. In the Mymensingh district, samples were collected from the Bangladesh Agricultural University campus area, Churkhai and Sutiakhali areas. In the Kushtia district samples were collected from Astanagar and Jhaudia. The water samples were initially processed, as indicated below, at the collection sites and then brought to the laboratory at the Department of Parasitology, Bangladesh Agricultural University, Mymensingh. Part of the work was done at the laboratory of Department of Microbiology and Hygiene, Bangladesh Agricultural University Mymensingh.
Sample collection and processing
During the present study, water samples were collected from various prospective waters sources like small ponds, waste water bodies and drains connected either directly or indirectly with animal houses, farms or grazing land. A total of 30 water samples were collected, 20 and 10 from the Mymensingh and Kushtia districts, respectively. For each sample 10 grab water samples from different location were collected in plastic jars and each grab sample was 10 liters. The water samples were initially passed through a mesh sieve (40mm pore size) to remove course particle and then 1 micron polyester filter bag (Duda, LLC, USA). The concentrated sample retained in the 1 micron filter bag was collected after mixing with 1% Tween 20 in a 50 ml tube. The samples were then centrifuged at 1500 x g for 30 min twice with distilled water for washing and again concentrated by the flotation technique using saturated salt solution (Najdrowski et al., 2007). A drop of the concentrate was smeared on a slide, stained by the modified Ziehl-Neelsen technique (Henriksen SA and Pohlenz, 1981) and examined under microscope (40X, 100X). The positive samples were washed with distilled water and stored at 4°C until use for DNA extraction.
Genomic DNA extraction
Oocysts were lysed after several cycles of freeze thawing, and DNA was extracted using Wizard genomic DNA purification kit (Promega, Madison, USA) according to the manufacturer’s instructions. Purified DNA was resuspended in a final volume of 25 µl of provided DNA rehydration buffer and stored at 4° C until use.
Polymerase chain reaction (PCR)
A primary PCR was performed by targeting the SSU rRNA gene of Cryptosporidium to confirm the detection of Cryptosporidium from water samples according to the protocol described by (Xiao et al., 1999) with some modification. A PCR product of 1,325 bp from target gene was amplified by forward (5ˊ-TTCTAGAGCTAATACATGCG- 3ˊ) and reverse primer (5ˊ- CCCATTTCCTTCGAAACAGGA -3ˊ) (IDT, USA). PCR reaction was performed in a total 50 µl reaction volume containing PCR Master mix (Promega, USA), 200 nM of each forward and reverse primer and 2 µl DNA sample. A total of 35 cycles were carried out, each consisting of 94° C for 45s, 53°C for 45s and 72°C for 1 min, with an initial hot start at 94°C for 3 min and a final extension at 72°C for 7 min. One kb DNA ladder (Promega, USA) was used to detect the band size of target DNA.
Agarose gel electrophoresis
PCR products were analyzed in 1.5% agarose gel, stained with ethidium bromide and examined against UV light using an image documentation system. The positive sample was recorded based on the appearance of expected size of band in the gel.
RESULTS AND DISCUSSION
Microscopic detection of Cryptosporidium
Conventional methods include examination of fecal smears with acid-fast stains such as Ziehl-Neelsen (Scott, 1988), which is commonly used by diagnostic facilities. Conventional microscopy is time-consuming and tedious and requires experienced person to accurately identify the oocysts (Garcia et al., 1987; Kehl et al., 1995) but still in use in many laboratories of the world as a cost-effective detection method of Cryptosporidium. In this study, Cryptosporidium oocyst from concentrated water sample was examined under the microscope following stained with Ziehl-Neelsen stain for initial screening of the sample before going to PCR steps.
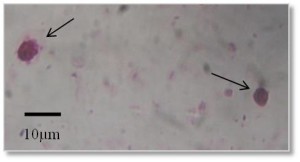
Oval or round shaped pink colored oocysts of Cryptosporidum were observed under microscope with pink background (Figure 1, 100X). In Mymensingh district, Cryptosporidium was found in 30% pond water samples and 40% waste water samples whereas the parasite was not detected in any samples collected from drain (Table 1).
Figure 1
Oocyst of Cryptosporidium (100X) stained with Ziehl Neelsen stain. Arrows indicate oocysts of Cryptosporidium. Scale bar: 10 μm.
Table 1
Detection of Cryptosporidium by microscopy with Ziehl Neelsen staining of samples collected at the Mymensingh district.
| Name of Samples | No of sample | No of positive sample | Percent positive sample |
| Pond water sample | 10 | 3 | 30 |
| Drain water Sample | 5 | 0 | 0 |
| Waste water sample | 5 | 2 | 40 |
| Total | 20 | 5 | 25 |
Sample were defined as positive by observing the presence of at least 2 oocysts of Cryptosporidium
Of the water sample from Kushtia district, 25% pond water samples and 33.33% drain water samples were Cryptosporidium positive whereas no waste water samples were positive for Cryptosporidium (Table 2).
Table 2
Detection of Cryptosporidium by microscopy with Ziehl Neelsen staining of samples collected at the Kushtia district.
| Name of Samples | No of sample | No of positive sample | Percent positive sample |
| Pond sample | 4 | 1 | 25 |
| Drain sample | 3 | 1 | 33.33 |
| Waste water sample | 3 | 0 | 0 |
| Total | 10 | 2 | 20 |
Sample were defined as positive by observing the presence of at least 2 oocysts of Cryptosporidium
The overall prevalence of Cryptosporidium in water samples was 20% in this study. In the Mymensingh district, the highest prevalence was recorded in waste water samples. This might be due to contamination of waste water with manure from Cryptosporidium-infected wild or farm animals. Cryptosporidium was not detected in drain water of the Mymensingh district. Probably the flow of drain water might take away the oocysts to the big reservoir. On the other hand, in the Kushtia district the higher prevalence of Cryptosporidium was observed in drain water sample compared to pond water samples might be due to the contamination of drain water with infected animals or farms and oocyst were retained the water bodies at the collection site.
The findings of this study conform to those of Hassan et al., (2012) who found Cryptosporidium in 27.5% water samples by microscopy. Cryptosporidium spp. infection was most common in children (44, 96%) (Khan et al., 2004). In that study, human infections were found in 7(25.9%) of 27 people examined. Cattle cryptosporidiosis cases was recorded in 7 (41.2%) of 17 examined, and goat cases in 3 (42.9%) of 7 examined (Park et al., 2006).
PCR detection of Cryptosporidium
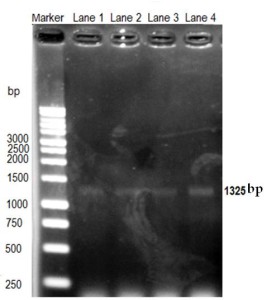
PCR-based methods have been used increasingly in the detection and analysis of Cryptosporidium oocysts in water (Xiao et al., 2004). The detection specificity and sensitivity is high in PCR based methods. The SSU rRNA gene was the target since this gene provides more than 99% accuracy to identify Cryptosporidum from different animal or host (Cai et al., 2001). The primers were selected to amplify a target sequence of 1325 bp size (Xiao et al., 1999). However, before examining the field sample, the PCR based reaction was optimized by using known DNA of Cryptosporidium. Different concentration of DNA was used and 100 ng DNA was found suitable to get optimum band of target DNA of Cryptosporidium.
The PCR reaction was performed by using different concentrations (12.5 ng- 100 ng) of DNA harvested from tentatively identified samples. A very clear thick band was detected during agarose gel electrophoresis using 100 ng of DNA sample, suggesting that 100 ng of DNA was required to get clear convencing band. Band was appeared at the expected size (1325bp) confirming the oocysts which was tentatively diagnosed by microscopy (Figure 2).
Figure 2
Optimization of Cryptosporidium detection by PCR. DNA concentration (12.5-100 ng) of sample was used for amplification. Lane 1= 12.5 ng; lane 2= 25 ng; lane 3= 50 ng; and lane 4= 100 ng DNA. Cryptosporidium target DNA band size is 1325 bp.
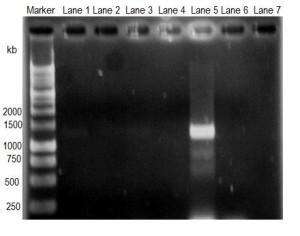
Figure 3
Amplification of Cryptosporidium target DNA obtained from water samples by PCR. Lane 1, pond water of Mymensingh; lane 2, pond water of Kushtia; lane 3, drain water of Mymensingh; lane 4, drain water of Kushtia; lane 5, waste water of Mymensingh; lane 6, waste water of Kushtia; and lane 7, negative control.
Only the positive samples of each category and all negative samples of each category were pooled in separate tube and subjected to DNA extraction. The DNA was subjected to PCR reaction to amplify target DNA of Cryptosporidium (1325bp). The gel image of resolved DNA in agarose demonstrated that DNA from pond water samples of Mymensingh (Figure 3, Lane 1) showed faint band whereas DNA from pond water samples of Kushtia (Figure 3, lane 2) did not show any visible band. DNA from drain water samples of Mymensingh (Figure 3, lane 3) showed faint band whereas DNA from drain water sample of Kushtia did not showed any band (Figure 3, lane 4). DNA from waste water sample of Mymensingh (Figure 3, lane 5) showed very strong band of 1325 kb, but DNA from waste water sample of Kushtia (Figure 3, lane 6) did not show any band.
In this study, the positive samples were pooled to increase the oocyst concentration as well as the amount of DNA concentration to increase the chance of detection of Cryptosporidium. The negative samples by microscopy were also pooled and subjected to DNA extraction if there is any oocyst present which could miss during microscopic examination.
The PCR detection of Cryptosporidium was found positive from most of the microscopically positive samples. The negative samples by microscopy were also negative by PCR detection methods. The pooled pond water and drain water samples from the Kushtia district were negative by PCR but were positive by microscopy. One possible explanation is the difficulty to discriminate between Cryptosporidium oocysts and Microsporidia by microscopy, as both of the organisms are very similar in size and both are acid fast. A similar observation was made by (Xiao et al., 2006) where six of the 24 microscopy-positive samples were negative by PCR.
In this study, PCR detection of Cryptosporidium oocysts in drain water of the Kushtia district was positive but was negative by microscopy probably due to higher sensitivity of PCR methods compared to traditional microscopy methods. However, low signals of some sample were observed which may be due to low concentration of Cryptosporidium DNA in total extracted DNA samples. In a sample of Mymensingh lane 5 (DNA from waste water sample) strong band of target sequence was observed probably the oocyst concentration as well amount of Cryptosporidium DNA in total extracted DNA was higher than the others.
Collectively, the results confirm the presence of Cryptosporidium in different types of sources of water in both Mymensingh and Kushtia district, indicating the existence or prevalence of Cryptosporidium in the study areas. However further studies are needed to explore the present status of prevalence of Cryptosporidium in humans and animals in order to better understanding the transmission dynamics of the parasite and thereafter taking necessary measure to control and prevent the disease.
ACKNOWLEDGEMENT
This work was supported by research Grants from the Swedish Research Council- 348-2012-6127, Sweden.
REFERENCES
Albert MJ, Faruque AS, Faruque SM, Sack RB and Mahalanabis D (1999). Case-control study of enteropathogens associated with childhood diarrhea in Dhaka, Bangladesh. Journal of Clinical Microbiology, 37: 3458–3464.
Alles AJ, Waldron MA, Sierra LS and Mattia AR (1995). Prospective comparison of direct immunofluorescence and conventional staining methods for detection of Giardia and Cryptosporidium in human fecal specimens. Journal of Clinical Microbiology, 33: 1632–1634.
Alves M, Matos O, Fonseca I, Delgado E, Lourenco A and Antunes F (2001). Multilocus genotyping of Cryptosporidium isolates from human HIV-infected and animal hosts. Journal of Eukaryotic Microbiology, Suppl 17S–18S.
Bhattacharya MK, Teka T, Faruque AS and Fuchs GJ (1997). Cryptosporidium infection in children in urban Bangladesh. Journal of Tropical Pediatrics, 43: 282–286.
Cai J, Collins MD, McDonald V, Thompson DE (2001). U.S. Environmental Protection Agency. Method 1623: Cryptosporidium and Giardia in water by filtration/IMS/FA. EPA 821-R-01-025. Office of Water, U.S. Environmental Protection Agency, Washington D.C.
Current W, Reese N, Ernst J, Bailey W, Heyman M and Weinstein W (1983). Human cryptosporidiosis in immunocompetent and immunodeficient persons. Studies of an outbreak and experimental transmission. The New England Journal of Medicine, 308: 1252– 1257.
Dalle F, Roz P, Dautin G, Di-Palma M, Kohli E, Sire-Bidault C, Fleischmann M G, Gallay A, Carbonel S, Bon F, Tillier C, Beaudeau P and Bonnin A (2003). Molecular characterization of isolates of waterborne Cryptosporidium spp. collected during an outbreak of gastroenteritis in South Burgundy, France. Journal of Clinical Microbiology, 41: 2690–2693.
Garcia LS, Brewer TC and Bruckner DA (1987). Fluorescence detection of Cryptosporidium oocysts in human fecal specimens using monoclonal antibodies. Journal of Clinical Microbiology, 25: 119–121.
Haque R, Mondal D, Kirkpatrick BD, Akther S, Farr BM, Sack RB and Petri WA Jr (2003). Epidemiologic and clinical characteristics of acute diarrhea with emphasis on Entamoeba histolytica infections in preschool children in an urban slum of Dhaka, Bangladesh. American Journal of Tropical Medicine and Hygiene, 69: 398-405.
Hassan A, Farouk H, Ghani RA and Hassanein F (2012). Contamination of irrigation systems of dental units with Cryptosporidium species in Alexandria, Egypt: a neglected disinfection pitfall. Risk Management and Healthcare Policy, 5: 93-95.
Henriksen SA and Pohlenz JF (1981). Staining of cryptosporidia by a modified Ziehl-Neelsen technique. Acta Veterinaria Scandinavica, 22: 594–596.
Kehl KSC, Cicirello H, Havens PL 1995: Comparison of four different methods for the detection of Cryptosporidium species. Journal of Clinical Microbiology, 33: 416–418.
Khan WA, Rogers KA, Karim MM, Ahmed S, Hibberd PL, Calderwood SB, Ryan ET and Ward HD (2004). Cryptosporidiosis among Bangladeshi children with diarrhea: a prospective, matched, case-control study of clinical features, epidemiology and systemic antibody responses. American Journal of Tropical Medicine and Hygiene, 71: 412-419.
Lake I, Pearce J and Savill M 2008: The seasonality ofhuman cryptosporidiosis in New Zealand. Epidemiology and Infection, Edinburgh Research Explorer, The University of Edinburgh, 136: 1383–1387.
Leoni F, Mallon ME, Smith HV, Tait A, McLauchlin J 2007: Multilocus analysis of Cryptosporidium hominis and Cryptosporidium parvum from sporadic and outbreak-related human cases and C. Parvum from sporadic cases in livestock in the UK. Journal of Clinical Microbiology, 45: 3286–3294.
MacKenzie WR, Hoxie NJ, Proctor ME, Gradus S, Blair KA, Peterson DE, Kazmierczak JJ, Addiss DG, Fox KR, Rose JP, Davis JP 1994: A massive outbreak in Milwaukee of Cryptosporidium infection transmitted through the public water supply. The New England Journal of Medicine, 331: 161–167.
McDonald V. Cryptosporidiosis. In: Cox FEG, (editor) 1996: The Welcome Trust illustrated history of tropical diseases. London, England: The Welcome Trust. pp. 256–263.
Mohandas K, Sehgal R, Sud A and Malla N (2002). Prevalence of intestinal parasitic pathogens in HIV-seropositive individuals in Northern India. Japanese Journal of Infectious Diseases, 55: 83–84.
Najdrowski M, Heckeroth AR, Wackwitz, C, Gawlowska S, Mackenstedt U, Kliemt D, Daugschies A (2007). Development and validation of a cell culture based assay for in vitro assessment of anticryptosporidial compounds. Parasitology Research, 101: 161–167.
O’Donoghue PJ 1995: Cryptosporidium and cryptosporidiosis in man and animals. International Journal for Parasitology, 25: 139–195.
Park JH, Guk SM, Han ET, Shin EH, Kim JL and Chai JY (2006). Genotype analysis of Cryptosporidium spp. prevalent in a rural village in Hwasun-gun, Republic of Korea. The Korean Journal of Parasitology, 44: 27-33.
Rahman AS, Sanyal SC, Al-Mahmud KA, Sobhan A 1985: Cryptosporidium diarrhoea in calves and their handlers in Bangladesh. The Indian Journal of Medical Research, 82: 510–516.
Rahman M, Shahid NS, Rahman H, Sack DA, Rahman N and Hossain S (1990). Cryptosporidiosis: a cause of diarrhea in Bangladesh. American Journal of Tropical Medicine and Hygiene, 42: 127-30.
Scott CD 1988: Screening fecal smears for Cryptosporidium and Isospora belli using a modified Ziehl-Neelsen stain. Journal of Medical Laboratory Science, 9: 80–82.
Tzipori S and Ward H (2002). Cryptosporidiosis: biology, pathogenesis and disease. Microbes and Infection; A Journal on Infectious Agents and Host Defenses, 4: 1047–1058.
WHO (2004). Guidelines for drinking water quality. 3rd edition. WHO, Geneva, Switzerland.
Xiao L Alderisio KA and Jiang J (2006). Detection of Cryptosporidium oocysts in water: effect of the number of samples and analytic replicates on test results. Applied and Environmental Microbiology, 72: 5942-7.
Xiao L, Bern C, Limor J, Sulaiman I, Roberts J, Checkley W, Cabrera L, Gilman RH and Lal AA (2001). Identification of 5 types of Cryptosporidium parasites in children in Lima, Peru. Journal of Infectious Diseases, 183: 492–497.
Xiao L, Lal AA and Jiang J (2004). Detection and differentiation of Cryptosporidium oocysts in water by PCR-RFLP. Methods in Molecular Biology, 268: 163-176.
Xiao L, Morgan UM, Limor J, Escalante A, Arrowood M, Shulaw W, Thompson RC, Fayer R and Lal AA (1999). Genetic diversity within Cryptosporidium parvum and related Cryptosporidium species. Applied and Environmental Microbiology, 65: 3386–3391.
Xiao L, Morgan UNAM, limor J, Escalante A, Arrowood M, Shulaw WRCA, Thompson, Fayer R and Lal AA (1999). Genetic Diversity within Cryptosporidium parvum and Related Cryptosporidium Species. Applied and Environmental Microbiology, 65: 3386–3391.
Zintl A, Proctor AF, Read C, Dewaal T, Shanaghy N, Fanning S and Mulcahy G (2008). The prevalence of Cryptosporidium species and subtypes in human faecal samples in Ireland. Epidemiology and Infection, 12: 1–8.